There’s an old saying about a dollar waiting on a dime. In other words, a monumental endeavor may become stuck at some point if a key preliminary element remains unavailable. The saying could be applied to the monumental endeavor we call synthetic biology. At present, it is waiting on synthetic DNA—long, high-quality, reasonably priced, and “on demand” synthetic DNA. When will the waiting end?
It’s a safe bet that the waiting won’t end all at once or as the result of any one technology. But there are many reasons to think that the waiting may end sooner rather than later. These include the technological advances described in this article. They all represent different ways of replacing chemical DNA synthesis with enzymatic DNA synthesis.
Chemical DNA synthesis, which generally relies on phosphoramidite technology, is currently the gold standard. However, it does present difficulties when longer, more complex oligonucleotides are needed. Phosphoramidite technology is usually limited to producing DNA sequences of about 200 base pairs in length. For longer sequences, error correction procedures can become unwieldy, and yields can drop. Although shorter sequences can be assembled to produce longer sequences, the presence of secondary structures and repetitive elements can complicate assembly. In general, chemical DNA synthesis can be costly and time consuming, and it can produce hazardous waste.
Enzymatic DNA synthesis promises to resolve many of the difficulties encountered with chemical DNA synthesis. For example, it promises to sustainably produce more complex templates at either bench scale or commercial scale. Strategies include innovative terminal deoxynucleotidyl transferase (TdT)-based processes and novel, cell-free amplification techniques. Ultimately, enzymatic DNA synthesis may help meet the growing demand for synthetic DNA across a range of applications. Indeed, as some scientists have put it (Hughes, Ellington. Cold Spring Harb. Perspect. Biol. 2017; 9: a023812), enzymatic approaches are “putting the synthetic in synthetic biology.”
Harnessing TdT
At Ansa Biotechnologies, two of the company’s co-founders, Dan Lin-Arlow, PhD, and Sebastian Palluk, PhD, came from the world of computational biology. Lured by the promise of synthetic biology, they entered the wet lab, where they soon grew frustrated by how much time was required to obtain DNA constructs. Eventually, they sought ways to reduce this time. As Ansa’s CSO and CTO, respectively, they contributed to the development of an enzymatic DNA synthesis method.
Like other such methods, it leverages TdT, a template-independent polymerase that was first characterized in the 1960s, and that was first used in a solid-phase DNA synthesis technology in the 1980s. TdT adds bases indiscriminately to DNA molecules. The challenge is controlling the enzyme so that it adds exactly one base and then temporarily stops, so that a desired DNA sequence can be synthesized step by step.
“Instead of reengineering TdT to accept blocked nucleotides, we tether a single unblocked nucleotide to the TdT enzyme via a linker so that each enzyme has only a single nucleotide that it can add, and no more,” Lin-Arlow says. “By preloading the enzyme with a native-like nucleotide substrate, our reactions are extremely fast and go to completion. After the addition is completed, we add a different enzyme to cleave the linker, exposing the end of the growing DNA molecule for the next addition cycle.”
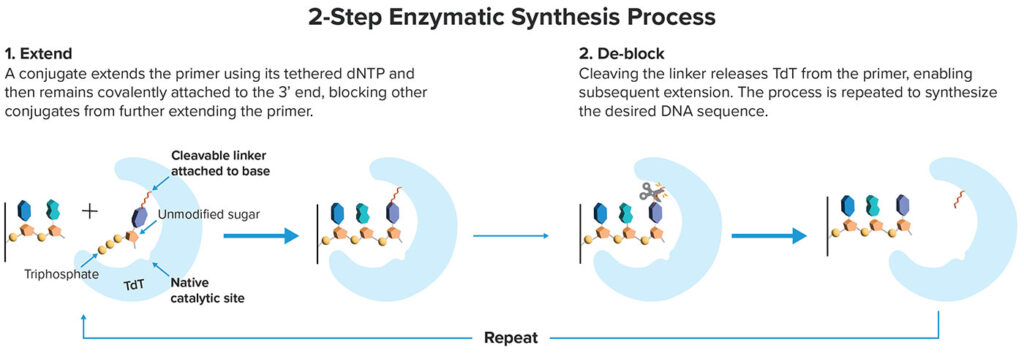
This simple and fast two-step cycle is repeated hundreds of times to synthesize the desired DNA sequence, including long oligos and complex sequences. “Scientists want what they ordered,” Lin-Arlow remarks. “They do not care how it is made.” To have a broader impact, enzymatic approaches have to produce higher-quality DNA than is possible with state-of-the-art chemical synthesis.
Scientists participating in Ansa’s expanding early access program can order DNA sequences of up to 600 base pairs, either in sequence-perfect clonal DNA format or a faster, sequence-verified linear double-stranded DNA fragment format. Sequences can include extreme GC content, tandem repeats, and secondary structures—features often found in genomic elements such as promoters, untranslated regions, and inverted terminal repeats.
“Our R&D team works hard to enhance the performance of our synthesis platform,” Lin-Arlow points out. “We anticipate being able to provide customers with even better DNA products going forward, including multikilobase DNA constructs in 2024.”
Evolving TdT
Core to Molecular Assemblies’ Fully Enzymatic Synthesis (FES) technology is the company’s TdT enzyme, which has had more than 25% of its amino acid sequence modified through a directed evolution process. According to the company, the modified TdT enzyme makes it possible for the FES technology to write DNA with cycle efficiencies of more than 99.9%, to deliver best-in-class purity, and to overcome any secondary structure challenges that arise from traditionally difficult sequences.
The process is a straightforward two-step synthesis cycle. The TdT enzyme incorporates a blocked nucleotide and precisely and accurately adds one base at a time. The blocking group is removed, and the process repeated to produce an oligo of the desired sequence and length.
The thermostable enzyme’s ability to operate efficiently and at extreme temperatures enables unique synthesis capabilities. “Customers are often limited by length, accuracy, and sequence complexity challenges,” says Phil Paik, PhD, CTO, Molecular Assemblies. “Our FES technology overcomes those limitations, enabling customers to unlock new research applications, such as in CRISPR therapeutics.”
The synthesis process is tunable from the femtomole scale, for gene assembly, to the picomole scale, for CRISPR and molecular cloning applications. And soon it will be tunable to the nanomole scale, for next-generation sequencing (NGS) and single-cell applications. The enzyme is capable of incorporating modified nucleotides and enables both single- and double-stranded products. Currently, single-stranded oligonucleotides up to 400 nt in length are available.
“Offering both a wide range of mass and complex sequences at length ensures we can provide synthesis capabilities in the size and flavor for a variety of research applications,” Paik asserts. “We have made sequences at extreme 100% GC or AT content, homopolymers of 250 nt-plus, and a variety of repetitive structures including inverted terminal repeats. We plan to expand our offering out toward 1 kb later this year.”
The company anticipates that its technology will contribute to a range of products based on the engineering of biology. Besides therapeutics and diagnostics, these products also include foods, cosmetics, and industrial chemicals. “At the heart of all these world-changing breakthroughs is the ability to manipulate biology’s programming language,” Paik observes. “DNA is an essential component and not simply a commodity.”
Benchtop synthesis using TdT
Over a decade ago, the founders of DNA Script envisioned that enzymatic synthesis of DNA would surpass chemical methods both in terms of performance and ease of use. Allowing scientists to “print” DNA directly on their own bench using a distributed model could transform biological science.
“Refining our Enzymatic DNA Synthesis (EDS) technology and building the Syntax instrument have been our core pillars,” says Rob Wilson, PhD, CCO, DNA Script. “Our TdT enzyme and our EDS technology can rapidly produce scarless, long oligonucleotides at a very high fidelity.”

Today, the Syntax technology allows scientists to work in their own laboratory and directly print DNA that is up to 120 nt long, with or without modifications, and scaled for multiple experiments. DNA Script has pushed its EDS technology even further in its R&D labs to synthesize highly pure and complex 400 nt long oligonucleotides on a routine basis. Early access to this improved biochemistry will come later this year.
The Syntax currently makes 120-mers at an industry-competitive error rate, and it can support traditionally difficult sequences (for example, those with high GC content or multiple repeats). Although the instrument is relatively new to the market, Wilson indicates that the company has seen good traction from pharmaceutical and genomics companies and contract research and development organizations that value IP security, control, convenience, and speed. Adoption is steadily increasing, and with upcoming improvements to system flexibility and usability, as well as to the EDS technology, DNA Script anticipates continued growth.
The next-generation Syntax will provide software upgrades and some hardware tweaks to improve the system’s flexibility. The company also plans to provide kits and reagents further enabling the EDS platform over the coming year.
“Oligonucleotide length, reduced error rate, complexity, and improved throughput are all key areas where customers will see benefits,” Wilson states. “By developing the best EDS technology and putting it into a single, self-contained Syntax instrument, we have demonstrated that we can successfully scale this new DNA synthesis approach.”
Cell-free cloning
According to Matthew Hill, PhD, founder and CEO of Elegen, perfect DNA cannot be synthesized directly. That is, fundamentally no chemistry, enzymatic or otherwise, will be good enough on its own to produce perfect DNA molecules directly, and thus cloning will always be required to isolate a perfect molecule from a population of imperfect ones.
In 1973, Stan Cohen and Herbert Boyer used a cell-based method for isolating and cloning perfect molecules. However, conventional cell-based cloning is challenging to scale. To overcome this difficulty, Elegen developed Enfinia DNA, a next-generation DNA manufacturing platform that permits a cell-free approach to cloning.

Besides selecting perfect molecules for replication, Enfinia DNA leverages automation to provide high-volume production from a bench-scale system. Furthermore, Hill points out, certain aspects of the process enable the production of DNA that is both longer and more complex than can be accomplished easily using cells. The customer-verified accuracy of Enfinia DNA is near-clonal.
“Our cell-free DNA manufacturing technology transforms ‘DNA Write’ the same way NGS transformed ‘DNA Read,’” Hill asserts. “It facilitates and speeds the acquisition of critical experimental inputs.”
Elegen ships NGS-verified, linear DNA, up to 7 kb, in six to eight days, and very complex DNA, including inverted terminal repeats, long terminal repeats, GC-rich elements, enhancers, and terminators as well as long homopolymers and repeats, in 10 to 15 business days. For example, a 14,713 bp plasmid containing a polyketide synthase gene cluster was delivered to the Keatinge-Clay laboratory at the University of Texas at Austin in 15 business days.
“Our recently announced collaboration with GSK validates the ability of our next-generation DNA production platform to supply critical material for the rapid discovery, development, and commercialization of genetic medicines,” Hill says. “It also reflects how we are expanding our offering for mRNA vaccines and cell and gene therapy programs. Our Series B financing will accelerate the extension to clinical DNA manufacturing.”
Long, complex ssDNA
Moligo Technologies focuses on the production of long, single-stranded DNA (ssDNA). These single-stranded oligonucleotides are important in research, in cell and gene therapy, in diagnostics probes, and in DNA nanotechnology. The company’s patented, PCR-free “injection molding” enzymatic technology can produce ultrapure DNA strands that are over 10,000 bases long and 100% sequence verified, and it can do so at scale. The process starts with a clonal template that is verified 100% correct, and then a polymerase with strand displacement activity directly produces ssDNA from these templates using rolling circle amplification in a scalable isothermal reaction. The company claims an error rate of 1 in 10 million bases.
Uses include gene therapy, in which nonviral genome targeting has many potential advantages, but some products, such as double-stranded DNA (dsDNA), pose challenges. The toxicity of naked dsDNA limits clinical applications with large nonviral DNA templates and has facilitated a movement toward the use of significantly less toxic ssDNA.
Moligo collaborates with Nationwide Children’s Hospital to provide long ssDNA for research into gene therapies for cystic fibrosis and other airway diseases. Using ssDNA to insert large genes, such as the cystic fibrosis transmembrane conductance regulator gene, opens up opportunities.
A cell therapy company, Cellectis, is evaluating Moligo’s long ssDNA donor templates in nuclease-mediated gene correction or gene insertion processes in long-term hematopoietic stem cells that are highly sensitive to the presence of foreign dsDNA. Cellectis and Moligo have already obtained results suggesting that the templates hold great promise for the field of ex vivo gene therapies. (Details were shared in a poster paper that a Cellectis scientist presented at the American Society of Gene and Cell Therapy conference this year.)
“To use DNA at large scale, you need a lot of high-quality material,” insists Cosimo Ducani, PhD, founder and CEO, Moligo. “We can produce, with high fidelity, sequences that include complexities such as repeats, high GC content, and secondary structure at industrial scale. We can also synthesize ssDNA molecules that are highly modified to have properties that are finely tuned for downstream applications.”
Error-free amplification
“More and more companies are finding ways to make complex DNA,” says Joe Hedley, PhD, CEO, NunaBio. “But unless you can make it at scale, its uses are limited. We are working on incorporating our amplification technology into our manufacturing process to create large quantities of DNA from scratch, very quickly, while maintaining fidelity and purity.”
A spinout of Newcastle University, NunaBio is developing technology that is based on work initially undertaken at Newcastle by Eimer Tuite, PhD, senior lecturer in biophysical chemistry, and Andrew Pike, PhD, senior lecturer in chemical nanoscience. The patented enzymatic amplification approach, not based on PCR or rolling circle amplification, can be used to synthesize cell-free DNA quickly.
NunaBio performs starting material synthesis, template assembly, and target amplification in house, and it has plans to internalize verification sequencing in the near future.

“Our customer-verified error rate is very low, 1 in 100,000 bases,” Hedley asserts. “This is because the amplification will not work if something goes wrong, for instance, if the wrong base is incorporated. We can provide complex materials including high GC content, sequences that have repetitive elements, modifications, as well as polyA tails up to 300 nucleotides. A new process is under development to add even longer polyA tails.”
The amplification technique is scalable using specially designed proprietary equipment that has the potential to produce large quantities of linear DNA in a very short time. To date, sequences up to 2–5 kb long have been produced.
According to Hedley, scientists usually reach out to NunaBio because they want a sequence they cannot get elsewhere, or because they have tried sequences unsuccessfully from other venders. Trial orders are available for new customers.
Customers range from big pharmaceutical companies to smaller biotechnology companies and research institutes. Products are varied. There are longer DNA templates with complex regions and polyA tails (for mRNA developers); shorter, simpler sequences (for antibody developers); and specific unique products (for researchers). The company is also investigating the possibility of providing, at scale, plasmids that incorporate the constructs.






